- Home
- Prototypes
- BioMedical Knowledge Extraction
BioMedical Knowledge Extraction
BeeSL: Towards industry-level biomedical event extraction
Biomedical event extraction has been a long-researched topic exhibiting pretty hard-to-tackle characteristics preventing existing solutions to exit the labs. The language understanding method described here represents the first option as a viable industrial solution to enhance the traditional pair-wise relation identification.
Events denote multiple, higher-order, associations among two or more interacting bio-entities describing, for example, changes on the state or location of the involved entities. The complexity of event extraction typically calls for multiple classifiers for recognizing event triggers and arguments.
As opposed to previous work, we followed a systems thinking approach to model all the sub-tasks in an end-to-end fashion, leading to a faster, joint model which also mitigates error propagation of locally-optimized classifier pipelines. We recast the task as a sequence labeling problem, proposing a novel multi-task deep neural network model with a BERT encoder pre-trained on biomedical texts, and soft-max and a novel multi-label classifier as decoder.
References
A. Ramponi, R. van der Goot, R. Lombardo, B. Plank. “Biomedical Event Extraction as Sequence Labeling”. Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP2020) link
A. Ramponi B. Plank, R. Lombardo, “Cross-Domain Evaluation of Edge Detection for Biomedical Event Extraction”. LREC, 2020 link
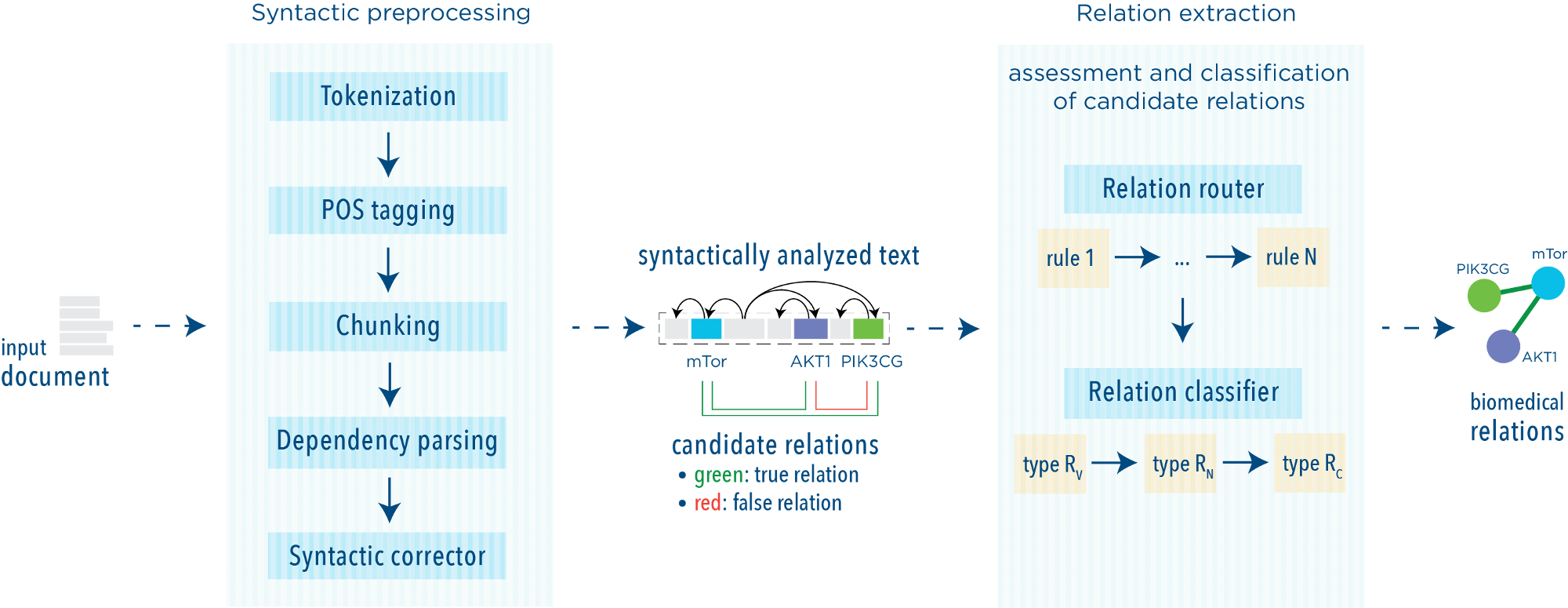
High-precision biomedical binary relation extraction system
The system is designed to extract highly precise relational information from input texts. In the figure we schematically show our approach to relation extraction, that includes two stages:
- text preprocessing, in which a sequence of natural language processing modules are applied to texts
- relation extraction, in which relationships between entities are identified and classified.
The system is available as a docker image, thus is platform independent.
It allows users to:
- extract entities in custom texts (providing input texts and input dictionaries);
- extract relations in custom texts (providing input texts and input dictionaries);
- replicate the results in gold standard corpora
Read install & run instructions, as well as benchmarks.
References
A. Ramponi, S. Giampiccolo, D. Tomasoni, C. Priami, and R. Lombardo, “High-Precision Biomedical Relation Extraction for Reducing Human Curation Efforts in Industrial Applications.” IEEE Access 8, 2169‑3536, 2020 link
R. Lombardo, S. Parolo, S. Michelini, L. Leonardelli, S. Giampiccolo, C. Kaddi, J. Barrett, K. Azer, “TB Knowledgebase: Interactive application for extracting knowledge from the TB literature to inform TB drug and vaccine development”. The 50th World Conference on Lung Health of the International Union Against Tuberculosis and Lung Disease (The Union), Volume: 23, 2019 link
S. Michelini, B. Balakrishnan, S. Parolo, A. Matone, J. Mullaney, W. Young, O. Gasser, C. Priami, R. Lombardo, M. Kussmann, “A reverse metabolic approach to weaning: In silico identification of immune-beneficial infant gut bacteria, mining their metabolism for prebiotic feeds and sourcing these feeds in the natural product space”. Microbiome, 2018 link